In this vignette we showcase how to use the cyCONDOR
ecosystem to predict cell type and cell state without manual annotation
of the dataset. This workflow is based on the Astir python
package, if you use this workflow please consider citing the Astir
manuscript Geuenich
et al. Cell Systems, 2021.
In cyCONDOR we use the reticulate package
to run python code from R. If you use the cyCONDOR
Docker image a conda environment is already configured to
run Astir. If you have a local installation of
cyCONDOR please visit Astir website to see a
tutorial on how to get the tool installed in your system.
Prepare your python environment
Conda load
If you are using cyCONDOR Docker container
you can visualize the list of available conda
environment.
## name python
## 1 base /opt/conda/bin/python
## 2 astir /opt/conda/envs/astir/bin/pythonActivate Conda
Now you simple need to activate Astir environment to be
ready to run this workflow.
use_condaenv(condaenv = "astir")Load example condor object
For this workflow we use an example dataset which was already
analysed with cyCONDOR.
condor <- readRDS("../.test_files/Astir/condor_example_astir.rds")Run Astir prediction
Astir allows to predict either cell type or cell state,
for more details on the package see the official manuscript (Geuenich
et al. Cell Systems, 2021).
For the prediction Astir needs a manifest
file where the characteristinc of each cell type or cell state are
specified.
This manifest file should be save as .yml
file with this structure:
## cell_types:
## CD4T:
## - CD3
## - CD4
## CD8T:
## - CD3
## - CD8
## NKT:
## - CD3
## - CD56
## NKBright:
## - CD56
## - CD16
## NKDim:
## - CD56
## B:
## - CD19
## pDCs:
## - CD123 (IL3RA)
## Classical_Monocytes:
## - CD14
## - HLA-DR
## cd16_Monocytes:
## - CD14
## - HLA-DR
## - CD16
##
## cell_states:
## Naive:
## - CD45RA
## Temra:
## - CD45RA
## - CD197 (CCR7)
## TCM:
## - CD197 (CCR7)You can now run the two functions for the prediction of the cell type
(run_astir_celltype) and cell state
(run_astir_cellstate).
Run Astrir to predict cell type
This function predict the cell type based on the marker selection
specified in the manifest file. The output of this function is saved
within the condor object under
condor$astir$Astir_cell_type_[data_slot]. Additionally some
QC data is saved in the analysis_path directory as
.csv
condor <- runAstir_celltype(fcd = condor,
data_slot = "orig",
analysis_path = "../.test_files/Astir/",
manifest_name = "marker.yml",
max_epochs = 1000,
learning_rate = 0.002,
initial_epochs = 3)## cell_type
## B cd16_Monocytes CD4T CD8T
## 1195 2171 17604 12338
## Classical_Monocytes NKBright NKDim NKT
## 13989 6589 669 1618
## Other pDCs Unknown
## 657 525 1694Run Astrir to predict cell state
Similarly to the previous function the
runAstir_cellstates function calculates a score for each
cell state declared in the manifest file. The results are saved in the
condor object in the slot
condor$astir$Astir_cell_state_[data_slot]. Also in this
case additional information are stored in the analysis_path
directory in .csv format.
condor <- runAstir_cellstates(fcd = condor,
data_slot = "orig",
analysis_path = "../.test_files/Astir/",
manifest_name = "marker.yml",
max_epochs = 1000,
learning_rate = 0.002,
initial_epochs = 3)Explore Astrir output
Cell type predictiom
## X cell_type
## 1 ID10.fcs_1 Classical_Monocytes
## 2 ID10.fcs_2 B
## 3 ID10.fcs_3 CD8T
## 4 ID10.fcs_4 CD8T
## 5 ID10.fcs_5 Classical_Monocytes
## 6 ID10.fcs_6 NKT## X CD4T CD8T NKT NKBright NKDim
## 1 ID10.fcs_1 1.367795e-07 1.040441e-06 2.237096e-08 2.666305e-08 2.577234e-09
## 2 ID10.fcs_2 1.324877e-09 1.897896e-08 8.172032e-09 1.478764e-06 8.104055e-07
## 3 ID10.fcs_3 7.930615e-07 9.996531e-01 3.249013e-04 2.514041e-11 9.783583e-07
## 4 ID10.fcs_4 4.006673e-06 9.991856e-01 8.099304e-04 1.315141e-08 1.246818e-08
## 5 ID10.fcs_5 4.936036e-08 4.064938e-08 3.321160e-09 1.654602e-09 1.129317e-08
## 6 ID10.fcs_6 4.400705e-04 1.701123e-03 9.978535e-01 6.876026e-07 1.317771e-09
## B pDCs Classical_Monocytes cd16_Monocytes Other
## 1 7.560338e-08 3.908119e-08 7.289595e-01 2.710360e-01 3.219714e-06
## 2 9.988234e-01 9.412180e-08 4.065692e-04 4.887747e-04 2.788651e-04
## 3 4.291974e-08 2.102051e-08 4.663446e-10 9.128537e-10 2.016069e-05
## 4 3.226020e-09 9.898312e-10 5.188650e-09 6.706455e-09 4.622255e-07
## 5 3.198307e-08 3.116859e-06 9.999630e-01 2.644898e-05 7.325300e-06
## 6 1.727692e-09 2.275862e-09 3.762259e-10 4.302245e-07 4.208238e-06Cell State
## X Naive Temra TCM
## 1 ID10.fcs_1 0.6374948 0.5499843 0.5300270
## 2 ID10.fcs_2 0.9017660 0.4957597 0.4049620
## 3 ID10.fcs_3 0.6665408 0.5347504 0.5063865
## 4 ID10.fcs_4 0.8504398 0.3773496 0.3034684
## 5 ID10.fcs_5 0.7882705 0.3863390 0.3289931
## 6 ID10.fcs_6 0.8909467 0.4584617 0.3715501Data Visualization
plot_dim_red(fcd = condor,
expr_slot = "orig",
reduction_method = "umap",
reduction_slot = "pca_orig",
add_astir = TRUE,
astir_slot = "Astir_cell_type_orig",
param = "cell_type",
order = T,
title = "UMAP colored by Astir predicted cell type",
facet_by_variable = FALSE,
raster = TRUE,
alpha = .3,
dot_size = .3)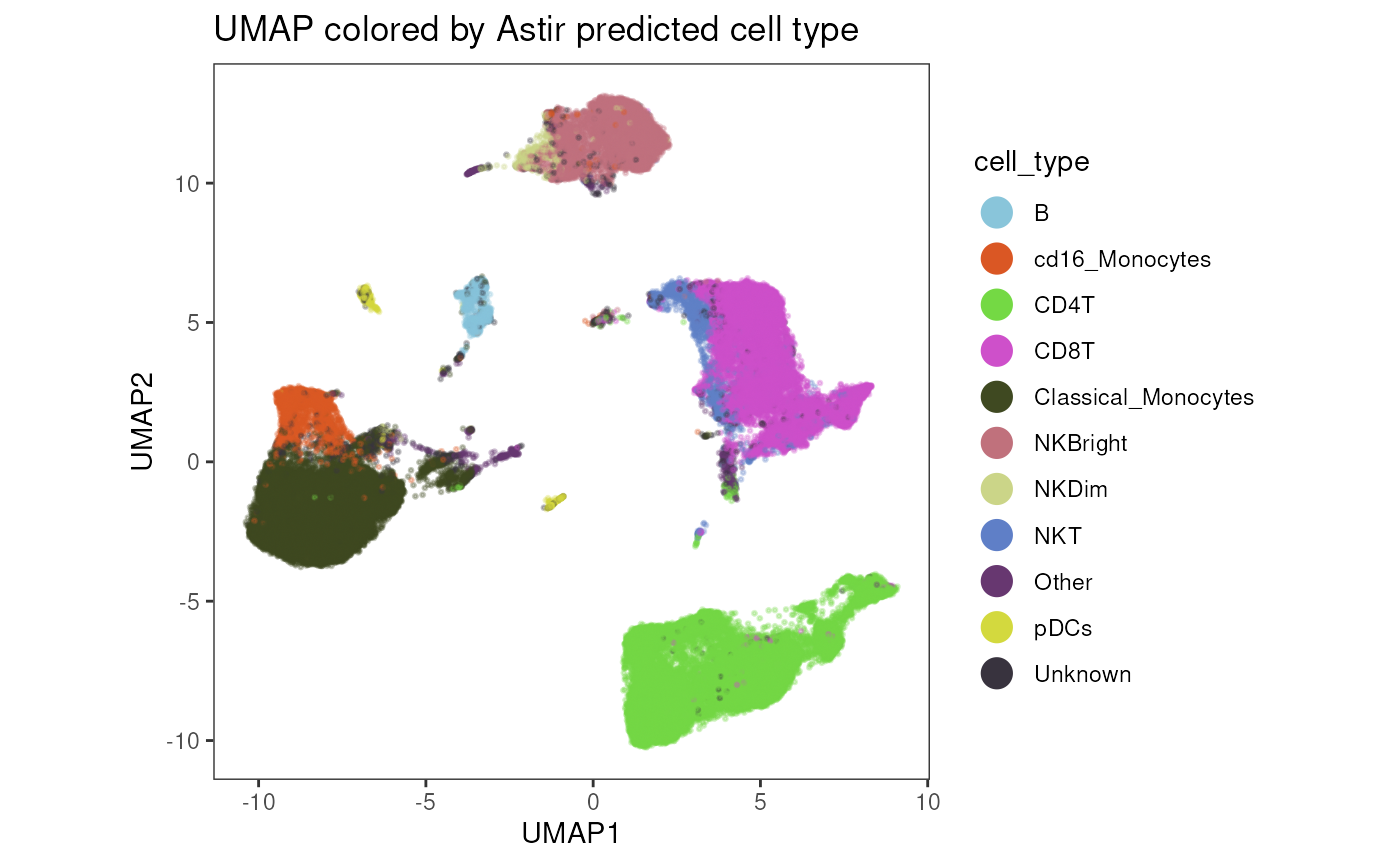
plot_dim_red(fcd = condor,
expr_slot = "orig",
reduction_method = "umap",
reduction_slot = "pca_orig",
add_astir = TRUE,
astir_slot = "Astir_cell_state_orig",
param = "Naive",
order = F,
title = "UMAP colored by Astir predicted cell state",
facet_by_variable = FALSE,
raster = TRUE,
alpha = .3,
dot_size = 1)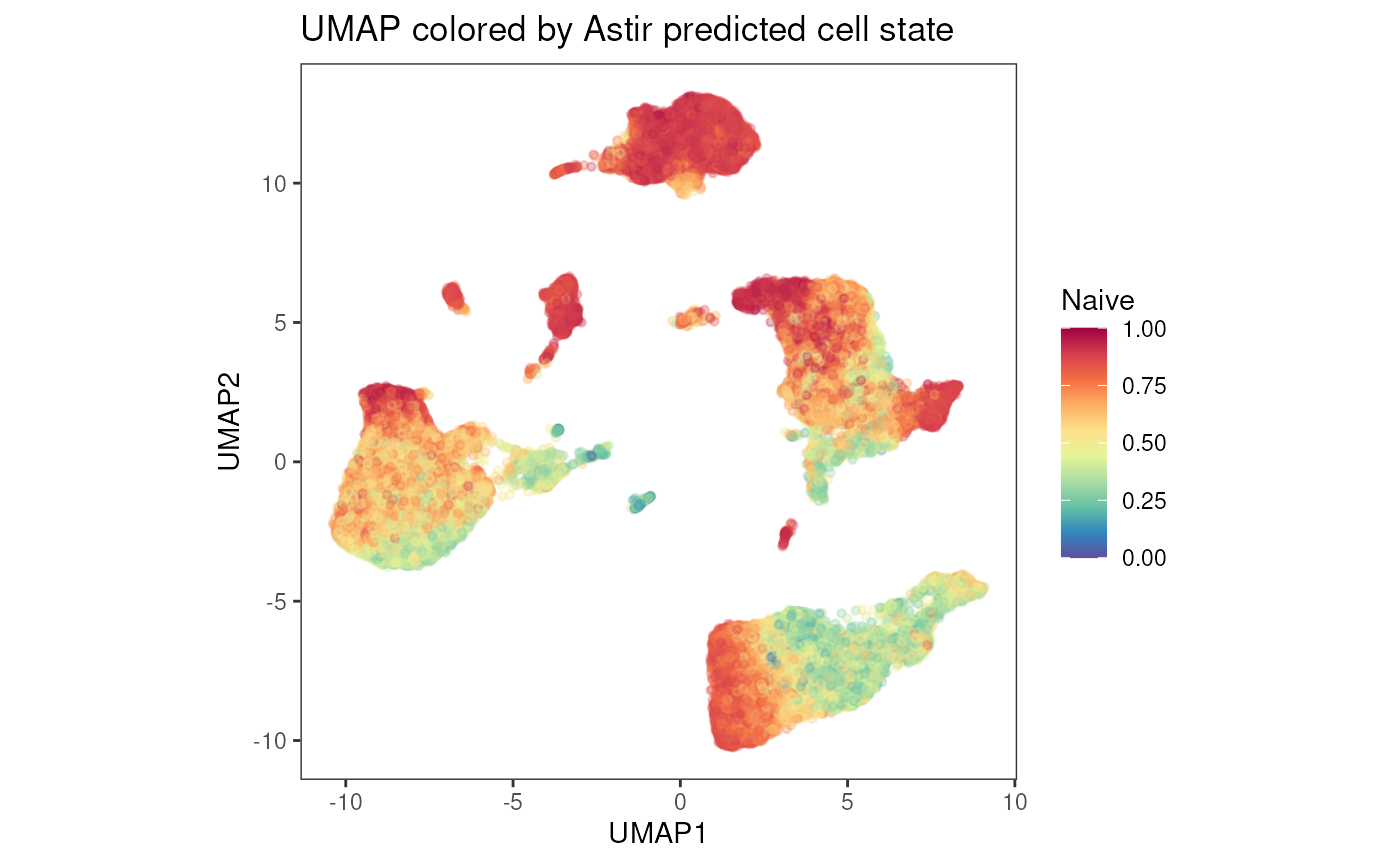
Session Info
info <- sessionInfo()
info## R version 4.4.2 (2024-10-31)
## Platform: x86_64-pc-linux-gnu
## Running under: Ubuntu 24.04.1 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## time zone: Etc/UTC
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] reticulate_1.42.0 cyCONDOR_0.3.1
##
## loaded via a namespace (and not attached):
## [1] IRanges_2.40.1 Rmisc_1.5.1
## [3] urlchecker_1.0.1 nnet_7.3-20
## [5] CytoNorm_2.0.1 TH.data_1.1-3
## [7] vctrs_0.6.5 digest_0.6.37
## [9] png_0.1-8 shape_1.4.6.1
## [11] proxy_0.4-27 slingshot_2.14.0
## [13] ggrepel_0.9.6 corrplot_0.95
## [15] parallelly_1.45.0 MASS_7.3-65
## [17] pkgdown_2.1.3 reshape2_1.4.4
## [19] httpuv_1.6.16 foreach_1.5.2
## [21] BiocGenerics_0.52.0 withr_3.0.2
## [23] ggrastr_1.0.2 xfun_0.52
## [25] ggpubr_0.6.1 ellipsis_0.3.2
## [27] survival_3.8-3 memoise_2.0.1
## [29] hexbin_1.28.5 ggbeeswarm_0.7.2
## [31] RProtoBufLib_2.18.0 princurve_2.1.6
## [33] profvis_0.4.0 ggsci_3.2.0
## [35] systemfonts_1.2.3 ragg_1.4.0
## [37] zoo_1.8-14 GlobalOptions_0.1.2
## [39] DEoptimR_1.1-3-1 Formula_1.2-5
## [41] promises_1.3.3 scatterplot3d_0.3-44
## [43] httr_1.4.7 rstatix_0.7.2
## [45] globals_0.18.0 rstudioapi_0.17.1
## [47] UCSC.utils_1.2.0 miniUI_0.1.2
## [49] generics_0.1.4 ggcyto_1.34.0
## [51] base64enc_0.1-3 curl_6.4.0
## [53] S4Vectors_0.44.0 zlibbioc_1.52.0
## [55] flowWorkspace_4.18.1 polyclip_1.10-7
## [57] randomForest_4.7-1.2 GenomeInfoDbData_1.2.13
## [59] SparseArray_1.6.2 RBGL_1.82.0
## [61] ncdfFlow_2.52.1 RcppEigen_0.3.4.0.2
## [63] xtable_1.8-4 stringr_1.5.1
## [65] desc_1.4.3 doParallel_1.0.17
## [67] evaluate_1.0.4 S4Arrays_1.6.0
## [69] hms_1.1.3 glmnet_4.1-9
## [71] GenomicRanges_1.58.0 irlba_2.3.5.1
## [73] colorspace_2.1-1 harmony_1.2.3
## [75] readxl_1.4.5 magrittr_2.0.3
## [77] lmtest_0.9-40 readr_2.1.5
## [79] Rgraphviz_2.50.0 later_1.4.2
## [81] lattice_0.22-7 future.apply_1.20.0
## [83] robustbase_0.99-4-1 XML_3.99-0.18
## [85] cowplot_1.2.0 matrixStats_1.5.0
## [87] xts_0.14.1 class_7.3-23
## [89] Hmisc_5.2-3 pillar_1.11.0
## [91] nlme_3.1-168 iterators_1.0.14
## [93] compiler_4.4.2 RSpectra_0.16-2
## [95] stringi_1.8.7 gower_1.0.2
## [97] minqa_1.2.8 SummarizedExperiment_1.36.0
## [99] lubridate_1.9.4 devtools_2.4.5
## [101] CytoML_2.18.3 plyr_1.8.9
## [103] crayon_1.5.3 abind_1.4-8
## [105] locfit_1.5-9.12 sp_2.2-0
## [107] sandwich_3.1-1 pcaMethods_1.98.0
## [109] dplyr_1.1.4 codetools_0.2-20
## [111] multcomp_1.4-28 textshaping_1.0.1
## [113] recipes_1.3.1 openssl_2.3.3
## [115] Rphenograph_0.99.1 TTR_0.24.4
## [117] bslib_0.9.0 e1071_1.7-16
## [119] destiny_3.20.0 GetoptLong_1.0.5
## [121] ggplot.multistats_1.0.1 mime_0.13
## [123] splines_4.4.2 circlize_0.4.16
## [125] Rcpp_1.1.0 sparseMatrixStats_1.18.0
## [127] cellranger_1.1.0 knitr_1.50
## [129] clue_0.3-66 lme4_1.1-37
## [131] fs_1.6.6 listenv_0.9.1
## [133] checkmate_2.3.2 DelayedMatrixStats_1.28.1
## [135] Rdpack_2.6.4 pkgbuild_1.4.8
## [137] ggsignif_0.6.4 tibble_3.3.0
## [139] Matrix_1.7-3 rpart.plot_3.1.2
## [141] statmod_1.5.0 tzdb_0.5.0
## [143] tweenr_2.0.3 pkgconfig_2.0.3
## [145] pheatmap_1.0.13 tools_4.4.2
## [147] cachem_1.1.0 rbibutils_2.3
## [149] smoother_1.3 fastmap_1.2.0
## [151] rmarkdown_2.29 scales_1.4.0
## [153] grid_4.4.2 usethis_3.1.0
## [155] broom_1.0.8 sass_0.4.10
## [157] graph_1.84.1 carData_3.0-5
## [159] RANN_2.6.2 rpart_4.1.24
## [161] farver_2.1.2 reformulas_0.4.1
## [163] yaml_2.3.10 MatrixGenerics_1.18.1
## [165] foreign_0.8-90 ggthemes_5.1.0
## [167] cli_3.6.5 purrr_1.1.0
## [169] stats4_4.4.2 lifecycle_1.0.4
## [171] uwot_0.2.3 askpass_1.2.1
## [173] caret_7.0-1 Biobase_2.66.0
## [175] mvtnorm_1.3-3 lava_1.8.1
## [177] sessioninfo_1.2.3 backports_1.5.0
## [179] cytolib_2.18.2 timechange_0.3.0
## [181] gtable_0.3.6 rjson_0.2.23
## [183] umap_0.2.10.0 ggridges_0.5.6
## [185] parallel_4.4.2 pROC_1.18.5
## [187] limma_3.62.2 jsonlite_2.0.0
## [189] edgeR_4.4.2 RcppHNSW_0.6.0
## [191] ggplot2_3.5.2 Rtsne_0.17
## [193] FlowSOM_2.14.0 ranger_0.17.0
## [195] flowCore_2.18.0 jquerylib_0.1.4
## [197] timeDate_4041.110 shiny_1.11.1
## [199] ConsensusClusterPlus_1.70.0 htmltools_0.5.8.1
## [201] diffcyt_1.26.1 rappdirs_0.3.3
## [203] glue_1.8.0 XVector_0.46.0
## [205] VIM_6.2.2 gridExtra_2.3
## [207] boot_1.3-31 TrajectoryUtils_1.14.0
## [209] igraph_2.1.4 R6_2.6.1
## [211] tidyr_1.3.1 SingleCellExperiment_1.28.1
## [213] labeling_0.4.3 vcd_1.4-13
## [215] cluster_2.1.8.1 pkgload_1.4.0
## [217] GenomeInfoDb_1.42.3 ipred_0.9-15
## [219] nloptr_2.2.1 DelayedArray_0.32.0
## [221] tidyselect_1.2.1 vipor_0.4.7
## [223] htmlTable_2.4.3 ggforce_0.5.0
## [225] CytoDx_1.26.0 car_3.1-3
## [227] future_1.58.0 ModelMetrics_1.2.2.2
## [229] laeken_0.5.3 data.table_1.17.8
## [231] htmlwidgets_1.6.4 ComplexHeatmap_2.22.0
## [233] RColorBrewer_1.1-3 rlang_1.1.6
## [235] remotes_2.5.0 colorRamps_2.3.4
## [237] Cairo_1.6-2 ggnewscale_0.5.2
## [239] hardhat_1.4.1 beeswarm_0.4.0
## [241] prodlim_2025.04.28